How To Implement Decision Tree Algorithms

Introduction
This beginner’s tutorial describes how to implement decision tree algorithms, namely random forests and gradient boosting machines. I’ll discuss how to train different models, search hyperparameters, and use ensembling techniques. To demonstrate this, I’ll use the scikit-learn library in Python and apply it to Kaggle’s Titanic competition, which has tabular data.
For tabular datasets (i.e., structured data like a spreadsheet), algorithms based on decision trees should be your first line of attack. These algorithms are fast and easy to implement (a GPU is not needed!), have high interpretability, and can produce similar results to deep neural networks.
In the Titanic competition, the goal is to accurately predict passenger survival. It’s a classification problem where performance will be measured using accuracy (i.e., the percentage of passengers who are correctly predicted).
Data Processing
The first step is to process the data.
For decision tree algorithms, it’s not necessary to perform one hot encoding to create dummy variables for ordered categorical variables. If they’re unordered, you’ll still need to create dummy variables. Why? Decision trees only care about the ordering of data, so it doesn’t matter whether your ordered categories are one-hot-encoded or not. For that same reason, it’s also not necessary to perform normalization or scaling of continuous variables.
Let’s get started by importing modules:
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
from kaggle import api
from sklearn.model_selection import cross_val_score, GridSearchCV, RandomizedSearchCV
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier, AdaBoostClassifier
from scipy.stats import randint
Specify some display options:
# Settings for matplotlib
%matplotlib inline
mpl.rc('axes', labelsize=14)
mpl.rc('xtick', labelsize=12)
mpl.rc('ytick', labelsize=12)
# Specify float format for pandas tables
pd.options.display.float_format = '{:.2f}'.format
Read in datasets to DataFrames. If you’re using a Kaggle notebook, the Titanic dataset will already be linked to your notebook. If you’re working locally, you can download the dataset to your filesystem.
train = pd.read_csv('../titanic/train.csv')
test = pd.read_csv('../titanic/test.csv')
List all of the columns, alongside their non-null count and data types:
train.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 891 entries, 0 to 890
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 PassengerId 891 non-null int64
1 Survived 891 non-null int64
2 Pclass 891 non-null int64
3 Name 891 non-null object
4 Sex 891 non-null object
5 Age 714 non-null float64
6 SibSp 891 non-null int64
7 Parch 891 non-null int64
8 Ticket 891 non-null object
9 Fare 891 non-null float64
10 Cabin 204 non-null object
11 Embarked 889 non-null object
dtypes: float64(2), int64(5), object(5)
memory usage: 83.7+ KB
This is what I know from the provided data dictionary:
Survivedis the column that I am trying to predict (1 = Survived, 0 = Died)Pclassis the ticket class (e.g., 1 for 1st class)SibSpis the number of siblings/spouses aboard the TitanicParchis the number of parents/children aboard the TitanicTicketis the ticket numberFareis the passenger fare amountCabinis the cabin numberEmbarkedis the port of embarkation (e.g., C for Cherbourg)
There are some missing values for Age, Cabin, and Embarked. In particular, Cabin is composed of mostly missing values, so I’m going to remove that column entirely.
I also like to look at the descriptive statistics for both object and number data types.
View statistics for object data types:
train.describe(include=object)
View statistics for number data types:
train.describe(include=np.number)
View columns with missing values, for both training and test sets:
train.isna().sum()
PassengerId 0
Survived 0
Pclass 0
Name 0
Sex 0
Age 177
SibSp 0
Parch 0
Ticket 0
Fare 0
Cabin 687
Embarked 2
dtype: int64
test.isna().sum()
PassengerId 0
Pclass 0
Name 0
Sex 0
Age 86
SibSp 0
Parch 0
Ticket 0
Fare 1
Cabin 327
Embarked 0
dtype: int64
The reason I looked at the missing values for the test set is because the training and test sets may differ in the columns that have missing values. As you can see above, the test set has a missing value in Fare that would have gone unnoticed if I was only looking at the training set.
I’ll perform median imputation for continuous variables Age and Fare, and mode imputation for the categorical variable Embarked.
Additionally, I want to remove Name and Ticket, since I don’t think those variables will be significant. I will also recode Sex to 1 (male) and 0 (female). Lastly, I will create dummy variables for Embarked because it’s unordered.
I’m going to summarize all of the action steps mentioned thus far in a single function, and apply it to the training data:
def process_data(df):
df = df.drop(labels=['Cabin', 'Name', 'Ticket'], axis=1)
df['Age'] = df['Age'].fillna(df['Age'].median())
df['Fare'] = df['Fare'].fillna(df['Fare'].median())
df['Embarked'] = df['Embarked'].fillna(df['Embarked'].mode().iloc[0])
df['Sex'] = df['Sex'].map({'male': 1, 'female': 0})
df = pd.get_dummies(df, columns=['Embarked'])
return df
train = process_data(train)
train.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 891 entries, 0 to 890
Data columns (total 11 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 PassengerId 891 non-null int64
1 Survived 891 non-null int64
2 Pclass 891 non-null int64
3 Sex 891 non-null int64
4 Age 891 non-null float64
5 SibSp 891 non-null int64
6 Parch 891 non-null int64
7 Fare 891 non-null float64
8 Embarked_C 891 non-null uint8
9 Embarked_Q 891 non-null uint8
10 Embarked_S 891 non-null uint8
dtypes: float64(2), int64(6), uint8(3)
memory usage: 58.4 KB
Separate the predictors from the labels, since they will be separately inputted to scikit-learn:
train_labels = train['Survived']
train = train.drop(labels='Survived', axis=1)
Apply the same data processing steps to the test set:
test = process_data(test)
And that’s it for data processing! Next, I’ll move on to training the model.
Model Training
Initial Approach
My initial approach is to quickly train a bunch of different models using default hyperparameter values to shortlist ones that look promising. At this stage, I don’t want to spend a lot of time training or tuning parameters before seeing results and submitting to a competition. In this case, I’m limiting myself to decision tree algorithms only, so there aren’t a lot of different ones to try.
In particular, I am interested in these specific models:
- decision tree
- random forest
- gradient boosting machine
To accomplish this, I’m going to write a function to loop through a list of scikit-learn models. During each loop, the model will be evaluated using scikit-learn’s cross_val_score, which performs k-fold cross validation.
Define a function to run the models and report their metrics:
def run_sklearn_models(list_of_models):
for m in list_of_models:
# Create an object for the model
sk_model = m
# Run k-fold cross-validation
scores = cross_val_score(sk_model, train, train_labels, scoring='accuracy', cv=10)
# Print results
print(m)
print(' Scores:', scores)
print(' Mean:', scores.mean())
print(' Standard Deviation:', scores.std())
run_sklearn_models([DecisionTreeClassifier(), RandomForestClassifier(), GradientBoostingClassifier()])
DecisionTreeClassifier()
Scores: [0.7 0.75280899 0.75280899 0.58426966 0.69662921 0.75280899
0.80898876 0.80898876 0.71910112 0.82022472]
Mean: 0.7396629213483145
Standard Deviation: 0.06679498865015357
RandomForestClassifier()
Scores: [0.82222222 0.80898876 0.75280899 0.79775281 0.82022472 0.80898876
0.78651685 0.79775281 0.84269663 0.82022472]
Mean: 0.8058177278401997
Standard Deviation: 0.023188209336299465
GradientBoostingClassifier()
Scores: [0.83333333 0.7752809 0.7752809 0.7752809 0.78651685 0.80898876
0.82022472 0.79775281 0.8988764 0.84269663]
Mean: 0.8114232209737828
Standard Deviation: 0.03730354000232365
As you can see, it looks like the GradientBoostingClassifier() is best, with RandomForestClassifier() a close second.
If you have other scikit-learn models you’re interested in trying, you can easily add their classes to the list accepted by the function run_sklearn_models().
At this point, you can proceed to the Inference & Submission section to create predictions for the test set using your best model and submit to Kaggle.
Hyperparameter Search
Now that I know which models look promising and I have a baseline level of performance from submitting to Kaggle, I’m going to invest extra time to search hyperparameter space to find values that result in the best performance.
I’m going to use scikit-learn’s RandomizedSearchCV to search through a distribution for each hyperparameter, and apply it to both RandomForestClassifier() and GradientBoostingClassifier().
Write a function to perform a hyperparameter search:
def random_search_models(m, n_iter, p):
# Run the RandomizedSearchCV()
print(m, '-------------------------')
model_object = m
rnd_search = RandomizedSearchCV(model_object, param_distributions=p, n_iter=n_iter,
cv=5, verbose=1, scoring='accuracy')
rnd_search.fit(train, train_labels)
# Save results to the DataFrame
df = pd.DataFrame.from_dict(rnd_search.cv_results_)
return df
Search hyperparameters for RandomForestClassifier(): (~15 minutes to run)
RFC_results = random_search_models(
RandomForestClassifier(), 500,
[{'n_estimators': randint(low=30, high=800),
'min_samples_leaf': randint(low=1, high=30),
'min_samples_split': randint(low=2, high=40),
'max_leaf_nodes': [None, 5, 10, 15, 20],
'max_depth': [None, 5, 10],
'bootstrap': [True, False],
'warm_start': [True, False]}])
RandomForestClassifier() -------------------------
Fitting 5 folds for each of 500 candidates, totalling 2500 fits
I chose a relatively large number of iterations (500). The parameter values are influenced by my past experience and from reading scikit-learn’s documentation for this model class.
The results are saved to a DataFrame so that I can easily refer to the accuracy and parameter values for each iteration.
View the columns of the resulting DataFrame:
RFC_results.columns
Index(['mean_fit_time', 'std_fit_time', 'mean_score_time', 'std_score_time',
'param_bootstrap', 'param_max_depth', 'param_max_leaf_nodes',
'param_min_samples_leaf', 'param_min_samples_split',
'param_n_estimators', 'param_warm_start', 'params', 'split0_test_score',
'split1_test_score', 'split2_test_score', 'split3_test_score',
'split4_test_score', 'mean_test_score', 'std_test_score',
'rank_test_score'],
dtype='object')
This DataFrame has recorded the parameter values that were chosen for each iteration as well as each split’s score (from k-fold cross validation) and their mean score.
Next, I’m going to plot accuracy vs. parameter in a scatter plot, where each point represents a specific iteration. This can be potentially useful since it’ll show which parameter values yield high accuracy.
Plot parameter vs. accuracy:
RFC_columns = ['param_bootstrap', 'param_max_depth', 'param_max_leaf_nodes',
'param_min_samples_leaf', 'param_min_samples_split',
'param_n_estimators', 'param_warm_start']
def plot_results(df, plot_columns):
for col in plot_columns:
fig, ax = plt.subplots()
ax.scatter(df[col], df['mean_test_score'])
plt.xlabel(col[6:])
plt.ylabel('accuracy')
plt.show()
plot_results(RFC_results, RFC_columns)
Here’s an example of a plot that showed a potential trend (min_samples_leaf parameter):
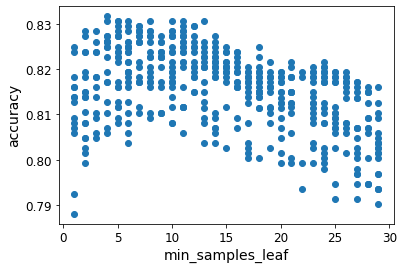
Here’s an example of a plot that didn’t show anything useful (min_samples_split parameter):
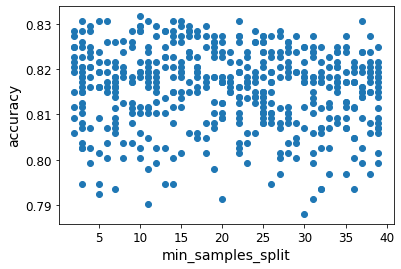
These plots did not end up being as illuminating as I’d hoped, since almost all plots showed no correlation.
I also searched hyperparameters for GradientBoostingClassifier(): (~10 minutes to run)
GBC_results = random_search_models(
GradientBoostingClassifier(), 500,
[{'learning_rate': [0.001, 0.003, 0.01, 0.03, 0.1, 0.3],
'n_estimators': randint(low=30, high=800),
'subsample': [0.1, 0.3, 0.7, 1.0],
'min_samples_leaf': randint(low=1, high=40),
'min_samples_split': randint(low=2, high=40),
'min_weight_fraction_leaf': [0.0, 0.3],
'max_depth': [3, 5, 10],
'warm_start': [True, False]}])
GradientBoostingClassifier() -------------------------
Fitting 5 folds for each of 500 candidates, totalling 2500 fits
Same as before: I plotted each parameter vs. accuracy, but did not see any correlations or trends in the figures.
View the top results from each run:
RFC_results.sort_values(by='mean_test_score', ascending=False).head(3)
GBC_results.sort_values(by='mean_test_score', ascending=False).head(3)
Both models have top results of about 83% accuracy on the validation set.
View the best-fit hyperparameters for each model:
RFC_results.sort_values(by='mean_test_score', ascending=False).head(1)['params'].squeeze()
{'bootstrap': False,
'max_depth': 10,
'max_leaf_nodes': 20,
'min_samples_leaf': 4,
'min_samples_split': 10,
'n_estimators': 45,
'warm_start': False}
GBC_results.sort_values(by='mean_test_score', ascending=False).head(1)['params'].squeeze()
{'learning_rate': 0.01,
'max_depth': 5,
'min_samples_leaf': 25,
'min_samples_split': 36,
'min_weight_fraction_leaf': 0.0,
'n_estimators': 449,
'subsample': 0.7,
'warm_start': True}
At this point, it’s a good idea to select your best model to generate predictions on the test set and submit them to the competition to see how you’re doing (see the Inference & Submission section).
Ensembling
One way that models can be further improved is by creating an ensemble of our best models, generate predictions on the test set from all of them, and select the most-voted prediction.
For example, create an ensemble from 5 different models:
submissions = test['PassengerId'].to_frame()
def run_ensemble(list_of_models):
for i, m in enumerate(list_of_models):
model_m = m
model_m.fit(train, train_labels)
predictions = model_m.predict(test)
submissions['Model' + str(i)] = predictions
run_ensemble([DecisionTreeClassifier(min_samples_leaf=5, min_samples_split=10, splitter='random'),
RandomForestClassifier(min_samples_leaf=5, n_estimators=400),
RandomForestClassifier(max_samples=0.8, min_samples_leaf=5, min_samples_split=15, n_estimators=400, warm_start=True),
GradientBoostingClassifier(),
GradientBoostingClassifier(learning_rate=0.01, n_estimators=200)])
Calculate the mode of their predictions:
submissions['Survived'] = submissions[['Model0', 'Model1', 'Model2', 'Model3', 'Model4']].mode(axis=1)
submissions.sample(n=20, random_state=5)
You can see how the models usually agree, but not always.
At this point, you can submit to the competition to see how well this technique worked in terms of improving your score.
Inference & Submission
Inference refers to generating predictions on the test set from your trained model. Submission refers to submitting a CSV file of your predictions to Kaggle. Ideally, you should be trying different techniques (such as those described in this blog post) frequently and submitting to Kaggle to gauge how well you’re doing.
First, train the model on the full training set using .fit:
best_model = GradientBoostingClassifier()
best_model.fit(train, train_labels)
Generate predictions on the test dataset using .predict:
predictions = best_model.predict(test)
submissions = test['PassengerId'].to_frame()
submissions['Survived'] = predictions
Save to a CSV file, since that’s what Kaggle expects:
submissions.to_csv('submission.csv', index=False)
Submit to Kaggle via the API:
!kaggle competitions submit -f submission.csv -m "Submission" -q "titanic"
Successfully submitted to Titanic - Machine Learning from Disaster
If you haven’t used Kaggle’s API yet, you can get started by reading the instructions on their Github.
And that’s it! You’ve successfully trained a model, generated predictions on the test set, and submitted them to Kaggle.
Leaderboard Score
Using the techniques described in this blog post, my best score was 78.7% and in the top 11% at the time of submission.
Most of my improvement came from simple hyperparameter tuning. Techniques that didn’t help were feature engineering and extensive hyperparameter tuning. It was unclear whether ensembling made a significant effect.
Resources
- Jeremy Howard’s Kaggle notebook on how random forests really work
- pandas library for data analysis and manipulation
- scikit-learn library for machine learning